lisTELOS
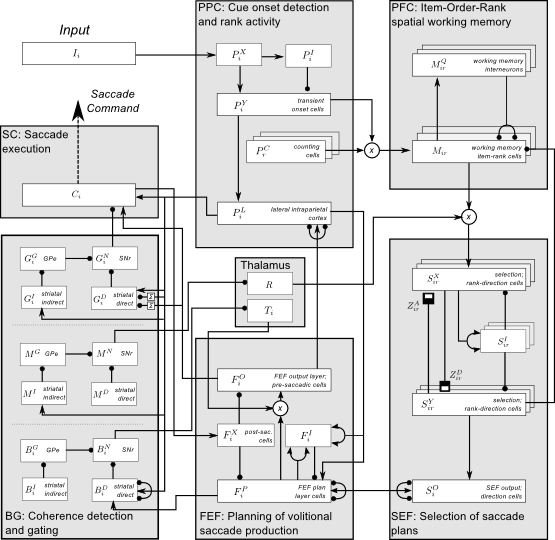
This code implements a neural model which is able to store sequences of spatial cues, even those with items repeated at arbitrary ordinal positions, and then produce a sequence of saccades to the cued locations once a central fixation point is extinguished.
In order to perform this and related tasks, the model exhibits a number of more fundamental properties including the ability to suppress reactive saccades toward cues presented during fixation, and the reproduction of reaction time data on benchmark saccade tasks (gap & overlap task, etc.).
Model simulations reproduce electrophysiological, psychophysical and behavioral data by successfully solving a number of oculomotor tasks. Particular emphasis is placed on the reproduction of data collected when the supplementary eye field is perturbed by cortical microstimulation. We hope to offer insight into the role of the supplementary eye field during the production of remembered saccade sequences by ensuring that the model's performance adheres to data from several manipulations of the region.
Requirements and installation
This code requires the only base installation of MATLAB. It has been fully tested on Linux systems, on which all simulations run smoothly. Tests in Windows systems seem to have been fine (but you never know) . Generally, the core model should work across platforms. Further tests are under way to ensure full compatibility with Windows.
No "installation" is necessary; simply put the lisTELOS folder somewhere on your computer.
First runs
Within MATLAB, navigate to the location of the 'installation' directory, called lisTELOS, and type:
[fields data] = simulateTrial;
this runs the model through a single trial of the immediate serial recall (ISR) task. The results of the simulation are passed back to the variable 'data', and the structure 'fields' contains information useful for accessing the data. Additional simulation information can be obtained through additional output arguments:
[fields data saccadeTimes saccadeTargets] = simulateTrial;
The two additional output arguments provide information about the times and targets, respectively, of saccades executed during the simulation.
The function simulateTrial accepts a number of optional arguments that control both the structure of the model and the task being simulated (see comments in simulateTrial for details). As an example, the simulation step size can be specified with the following command:
[fields data] = simulateTrial('Step', 0.0001);
To specify multiple values, use the standard method for specifying multiple arguments in MATLAB functions. For example:
[fields data] = simulateTrial('Step', 0.0001, 'StimLocation', 23);
Through these arguments (particularly the more powerful values such as CueLocations, CueOnTimes, and CueOffTimes), the structure of most basic behavioral tasks can be provided to the model. All simulations in the published paper ran trials through this method.
Notes on timing
When using the model, express timing-related parameters in seconds.
But read on...
A choice early in implementation led to the representation of simulation time in hundreds of milliseconds, rather than seconds. This doesn't impact the performance of the model -- or the accuracy with which it can resolve time -- it just means that a different time unit is used when performing simulations.
In simulations included in this package (see below) all time-related inputs to the model are expressed in seconds. Any new simulations should also express time in seconds. The first steps taken by the simulateTrial function convert times to the simulation units, and the last lines convert all times back to seconds. In addition, all text printed to the matlab console during simulations are expressed in seconds.
All this is to say: from the user's perspective, the model deals with time in seconds. Any modification to the model itself should be done with care, and with full knowledge of the shift in units.
Key simulations
To reproduce model simulations described in the paper, one can execute one of several task functions included in this package. These functions use the simulateTrial function, specifying task parameters as arguments, and then plot the results. In many cases, these functions reproduce figures from the paper exactly. The functions available, and corresponding figures in Silver et al. (2012), are:
| Task Function | Figure | Description |
|---|---|---|
| BenchmarkSaccadeTasks.m | 5 | Simulates four benchmark saccade tasks (Saccade, Overlap, Delayed Saccade, and Gap 500) and plots stimuli and eye movements for each. |
| SaccadeTask.m | 6 | Simulates the Saccade (Gap 0) task and plots selected cell traces. |
| OverlapTask.m | 7 | Simulates the Overlap task and plots selected cell traces. |
| DelayedSaccadeTask.m | 8 | Simulates the Delayed Saccade task and plots selected cell traces. |
| GapTask.m | 9 | Simulates the Gap 500 task and plots selected cell traces. |
| ImmediateSerialRecallTask.m | 11 | Simulates the Immediate Serial Recall (ISR) task with a four item sequence (containing a repeated item) and plots selected cell traces. |
| HistedMillerTask.m | 13 | Simulates the modified double saccade task used by Histed and Miller (2006) to show that SEF microstimulation can reorder a remembered spatial sequence. Selected cell traces are plotted. |
| HistedMillerBatch.m | 12, 15 | Simulates a batch of trials from the Histed and Miller (2006) double saccade task. This function produces data used to create Figure 12. Because of stochasticity in the simulations, results may vary significantly between simulations. Because this function takes long, the results are saved in a file called backupData.mat before plotting. Plots corresponding to Figure 15 are saved as eps files, and not drawn on the screen. This speeds things up when many figures are to be generated. |
| YangOthersTask.m | 16 | Simulates the task used by Yang, Heinen, and Missal (2008) to show that SEF microstimulation can, under some conditions, change saccade latency. |
If additional simulations are desired, it is highly recommended that users do so in the same fashion as the above functions. For help representing a task in a way that the simulateTrial function can accept, please do not hesitate to contact Matt Silver:
Contact
Thanks for taking the time to examine this model! Please write with any comments, concerns, criticisms, compliments, corrections, et cetera, to silverm@mit.edu